Gene regulatory regions drive cell type- and cell state-specific gene expression. However, while efforts to identify regions that are likely to regulate gene expression have been widely successful, the vast majority of these regions do not have a verified target gene (or genes) nor a context for their activity. Our goal is to map regulatory regions genome-wide in immune cells to better understand what genes they regulate, the context required for enhancer regulation and to computationally derive generalizable rules for their activity that can be applied to many other cell types.
Since autoimmune disease-associated genetic variants are enriched in immune cell regulatory regions, these regions are likely key in driving autoimmunity. Millions of putative regulatory regions have been mapped through assaying chromatin accessibility, histone modifications and 3-D chromatin conformation. However, despite the wealth of publicly available data for many cell types, we are still in the early stages of predicting how immune cell regulatory regions operate:
- Their strength and directionality on gene expression are still poorly predicted
- Connecting regulatory regions to the genes they regulate remains an extraordinary challenge
To address this, a major focus of the lab is to connect these regions to gene expression and cellular function at baseline and in activation states, with the goal of finding the ones that influence trait-relevant cellular states. One of our key tools is CRISPR-interference (CRISPRi), a catalytically dead Cas9 protein tethered to a KRAB domain. When targeted to an active regulatory region in the genome, CRISPRi silences its activity. We are currently using CRISPRi to determine variant enhancers that modulate immune cell function.
Additional Research Projects
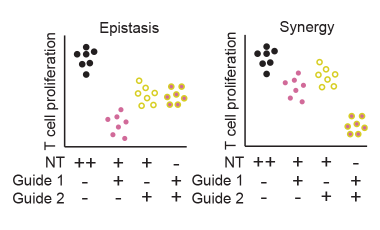
Defining variants that act collectively to promote disease
Identifying synergistic properties of variants and enhancers on T cell function
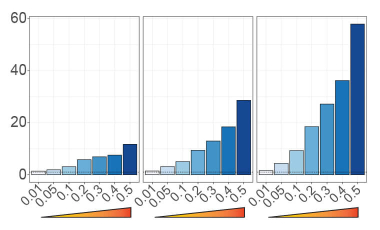
Prioritizing autoimmune disease-causal variants in immune cells
Utilizing massively parallel reporter assays and statistical fine-mapping to enrich for functional and disease-causal genetic variation



